Use baredSC within python¶
This part is not a proper documentation of baredSC as a package as we are missing a lot of documentation of our methods. It just gives indications on how it can be used.
baredSC_1d¶
Use the npz content¶
baredSC is mainly used with the command line interface.
By default, only the numpy compressed .npz file is output,
but if the --figure argument is used, it outputs much more.
All the outputs are described in the Outputs page.
We provide two examples of plots using the text outputs in Compare means from baredSC results and Customize your baredSC plots. However, sometimes the user may want to plot information which is not part of the text outputs. We describe here a script which will use the baredSC_1d output to plot a custom error bars.
First we get the value of parameters at each step of MCMC:
In [1]: import numpy as np
...: import baredSC.oned
...: import matplotlib.pyplot as plt
...:
...: output_baredSC = "../example/first_example_1d_2gauss.npz"
...:
...: # First option, use the function extract_from_npz
...: mu, cov, ox, oxpdf, x, logprob_values, samples = \
...: baredSC.oned.extract_from_npz(output_baredSC)
...: # mu is the mean of each parameter,
...: # cov is the covariance of parameters
...: # ox is the oversampled x to compute the poisson noise pdf
...: # oxpdf is the oversampled x to compute the pdf
...: # x is the x used to compute the likelihood
...: # logprob_values is the value of log likelihood at each step fo the MCMC
...: # samples is the value of each parameter at each step of the MCMC
...:
...: # Second option, get the samples and x, oxpdf from the npz:
...: mcmc_res = np.load(output_baredSC, allow_pickle=True)
...: samples = mcmc_res['samples']
...: x = mcmc_res['x']
...: oxpdf = mcmc_res['oxpdf']
...:
...: # From the sample size we deduce the number of Gaussians
...: nnorm = (samples.shape[1] + 1) // 3
...: # We display the parameter names:
...: p_names = [f'{pn}{i}' for i in range(nnorm) for pn in ['amp', 'mu', 'scale']][1:]
...: print(f'The parameters are: {p_names}')
...:
Reading
Read. It took 0.03 seconds.
The parameters are: ['mu0', 'scale0', 'amp1', 'mu1', 'scale1']
Then we compute the pdf for each step of the MCMC and we plot the pdf with the custom error bars:
In [2]: # We assume x is equally spaced
...: dx = x[1] - x[0]
...: nx = x.size
...: noxpdf = oxpdf.size
...: # We assume oxpdf is equally spaced
...: odxpdf = oxpdf[1] - oxpdf[0]
...:
...: # Compute the pdf for each sample
...: # This can be long
...: pdf = np.array([baredSC.oned.get_pdf(p, nx, noxpdf, oxpdf, odxpdf)
...: for p in samples])
...:
...: xmin = x[0] - dx / 2
...: xmax = x[-1] + dx / 2
...:
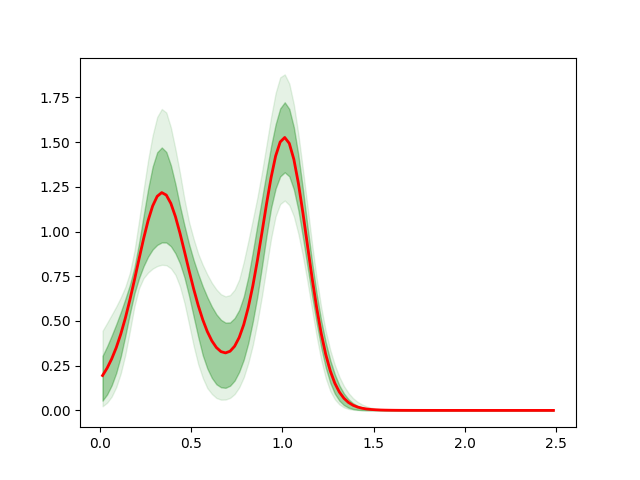
...: my_custom_quantiles = {'0.3': [0.25, 0.75], '0.1': [0.1, 0.9]}
...:
...: plt.figure()
...: for alpha in my_custom_quantiles:
...: pm = np.quantile(pdf, my_custom_quantiles[alpha][0], axis=0)
...: pp = np.quantile(pdf, my_custom_quantiles[alpha][1], axis=0)
...: plt.fill_between(x, pm, pp, color='g', alpha=float(alpha),
...: rasterized=True)
...: # Mean
...: plt.plot(x, np.mean(pdf, axis=0), 'r', lw=2, rasterized=True)
...:
Out[2]: [<matplotlib.lines.Line2D at 0x7f455406b590>]
Run baredSC_1d¶
You can also run the MCMC from python directly.
However, it requires formating of the input:
In [3]: import numpy as np
...: import pandas as pd
...: from scipy.stats import lognorm, truncnorm, poisson
...: from baredSC.baredSC_1d import gauss_mcmc
...:
...: # I generate 200 cells with normal expression at 1.5 with scale of 0.2
...: # In the Seurat scale (log(1 + 10^4 X))
...: n_cells = 200
...: cur_loc = 1.5
...: cur_scale = 0.2
...: N = lognorm.rvs(s=0.3, scale=16000, size=n_cells, random_state=1).astype(int)
...: expression = truncnorm.rvs(- cur_loc / cur_scale, np.inf,
...: loc=cur_loc, scale=cur_scale,
...: size=n_cells,
...: random_state=2)
...:
...: ks = poisson.rvs(mu=N * 1e-4 * (np.exp(expression) - 1),
...: random_state=3)
...:
...: # I need to put the ks and the N in a data frame:
...: # The column containing the total number of UMI per cell
...: # must be 'nCount_RNA'
...: data = pd.DataFrame({'my_gene': ks, 'nCount_RNA': N})
...:
Then the actual MCMC can be run with:
In [4]: results = gauss_mcmc(data=data,
...: col_gene='my_gene', # Put here the colname you put in your data
...: nx=50, # Number of bins in x
...: osampx=10, # Oversampling factor of the Poisson distribution
...: osampxpdf=5, # Oversampling factor of the PDF
...: xmin=0,
...: xmax=3,
...: min_scale=0.1, # Minimal value of the scale
...: xscale="Seurat",
...: target_sum=10000,
...: nnorm=1, # We use models with a single Gaussian
...: nsamples_mcmc=100000, # Number of steps in the MCMC
...: nsamples_burn=25000, # Number of steps in the burning phase of MCMC (we recommand nsampMCMC / 4)
...: nsplit_burn=10, # The burning phase is splitted in multiple sub-phase where the temperature is decreasing
...: T0_burn=100.0,
...: output='temp', # Where the npz output should be stored
...: seed=1)
...: print(f'results contains {len(results)} items.')
...: # The results are:
...: # mu, cov, ox, oxpdf, x, logprob_values, samples
...: # mu is the mean of each parameter,
...: # cov is the covariance of parameters
...: # ox is the oversampled x to compute the poisson noise pdf
...: # oxpdf is the oversampled x to compute the pdf
...: # x is the x used to compute the likelihood
...: # logprob_values is the value of log likelihood at each step fo the MCMC
...: # samples is the value of each parameter at each step of the MCMC
...:
Step 1000, acceptance rate (since last printing): 0.2330
Step 2000, acceptance rate (since last printing): 0.2580
Step 1000, acceptance rate (since last printing): 0.2400
Step 2000, acceptance rate (since last printing): 0.2370
Step 1000, acceptance rate (since last printing): 0.2450
Step 2000, acceptance rate (since last printing): 0.2430
Step 1000, acceptance rate (since last printing): 0.2350
Step 2000, acceptance rate (since last printing): 0.3930
Step 1000, acceptance rate (since last printing): 0.2580
Step 2000, acceptance rate (since last printing): 0.2060
Step 1000, acceptance rate (since last printing): 0.2680
Step 2000, acceptance rate (since last printing): 0.2440
Step 1000, acceptance rate (since last printing): 0.2640
Step 2000, acceptance rate (since last printing): 0.2440
Step 1000, acceptance rate (since last printing): 0.2350
Step 2000, acceptance rate (since last printing): 0.2400
Step 1000, acceptance rate (since last printing): 0.2450
Step 2000, acceptance rate (since last printing): 0.2340
Step 1000, acceptance rate (since last printing): 0.2530
Step 2000, acceptance rate (since last printing): 0.2460
Step 1000, acceptance rate (since last printing): 0.2060
Step 2000, acceptance rate (since last printing): 0.2330
Step 3000, acceptance rate (since last printing): 0.2470
Step 4000, acceptance rate (since last printing): 0.2410
Step 5000, acceptance rate (since last printing): 0.2360
Step 6000, acceptance rate (since last printing): 0.2380
Step 7000, acceptance rate (since last printing): 0.2210
Step 8000, acceptance rate (since last printing): 0.2050
Step 9000, acceptance rate (since last printing): 0.2500
Step 10000, acceptance rate (since last printing): 0.2620
Step 11000, acceptance rate (since last printing): 0.2240
Step 12000, acceptance rate (since last printing): 0.2310
Step 13000, acceptance rate (since last printing): 0.2350
Step 14000, acceptance rate (since last printing): 0.2480
Step 15000, acceptance rate (since last printing): 0.2360
Step 16000, acceptance rate (since last printing): 0.2400
Step 17000, acceptance rate (since last printing): 0.2190
Step 18000, acceptance rate (since last printing): 0.2260
Step 19000, acceptance rate (since last printing): 0.2360
Step 20000, acceptance rate (since last printing): 0.2350
Step 21000, acceptance rate (since last printing): 0.2380
Step 22000, acceptance rate (since last printing): 0.2100
Step 23000, acceptance rate (since last printing): 0.2500
Step 24000, acceptance rate (since last printing): 0.2510
Step 25000, acceptance rate (since last printing): 0.2620
Step 26000, acceptance rate (since last printing): 0.2480
Step 27000, acceptance rate (since last printing): 0.2610
Step 28000, acceptance rate (since last printing): 0.2530
Step 29000, acceptance rate (since last printing): 0.2180
Step 30000, acceptance rate (since last printing): 0.2410
Step 31000, acceptance rate (since last printing): 0.2340
Step 32000, acceptance rate (since last printing): 0.2170
Step 33000, acceptance rate (since last printing): 0.2290
Step 34000, acceptance rate (since last printing): 0.2440
Step 35000, acceptance rate (since last printing): 0.2550
Step 36000, acceptance rate (since last printing): 0.2360
Step 37000, acceptance rate (since last printing): 0.2060
Step 38000, acceptance rate (since last printing): 0.2360
Step 39000, acceptance rate (since last printing): 0.2630
Step 40000, acceptance rate (since last printing): 0.2340
Step 41000, acceptance rate (since last printing): 0.2150
Step 42000, acceptance rate (since last printing): 0.2460
Step 43000, acceptance rate (since last printing): 0.2600
Step 44000, acceptance rate (since last printing): 0.2210
Step 45000, acceptance rate (since last printing): 0.2310
Step 46000, acceptance rate (since last printing): 0.2350
Step 47000, acceptance rate (since last printing): 0.2210
Step 48000, acceptance rate (since last printing): 0.2280
Step 49000, acceptance rate (since last printing): 0.2480
Step 50000, acceptance rate (since last printing): 0.2200
Step 51000, acceptance rate (since last printing): 0.2370
Step 52000, acceptance rate (since last printing): 0.2270
Step 53000, acceptance rate (since last printing): 0.2590
Step 54000, acceptance rate (since last printing): 0.2170
Step 55000, acceptance rate (since last printing): 0.2240
Step 56000, acceptance rate (since last printing): 0.2530
Step 57000, acceptance rate (since last printing): 0.1990
Step 58000, acceptance rate (since last printing): 0.2370
Step 59000, acceptance rate (since last printing): 0.2200
Step 60000, acceptance rate (since last printing): 0.2560
Step 61000, acceptance rate (since last printing): 0.2360
Step 62000, acceptance rate (since last printing): 0.2290
Step 63000, acceptance rate (since last printing): 0.2600
Step 64000, acceptance rate (since last printing): 0.2060
Step 65000, acceptance rate (since last printing): 0.2260
Step 66000, acceptance rate (since last printing): 0.2550
Step 67000, acceptance rate (since last printing): 0.2470
Step 68000, acceptance rate (since last printing): 0.2410
Step 69000, acceptance rate (since last printing): 0.2490
Step 70000, acceptance rate (since last printing): 0.2450
Step 71000, acceptance rate (since last printing): 0.2160
Step 72000, acceptance rate (since last printing): 0.2130
Step 73000, acceptance rate (since last printing): 0.2340
Step 74000, acceptance rate (since last printing): 0.2410
Step 75000, acceptance rate (since last printing): 0.2290
Step 76000, acceptance rate (since last printing): 0.2240
Step 77000, acceptance rate (since last printing): 0.2350
Step 78000, acceptance rate (since last printing): 0.2460
Step 79000, acceptance rate (since last printing): 0.2280
Step 80000, acceptance rate (since last printing): 0.2580
Step 81000, acceptance rate (since last printing): 0.2530
Step 82000, acceptance rate (since last printing): 0.2240
Step 83000, acceptance rate (since last printing): 0.2490
Step 84000, acceptance rate (since last printing): 0.2190
Step 85000, acceptance rate (since last printing): 0.2350
Step 86000, acceptance rate (since last printing): 0.2160
Step 87000, acceptance rate (since last printing): 0.2480
Step 88000, acceptance rate (since last printing): 0.2460
Step 89000, acceptance rate (since last printing): 0.2430
Step 90000, acceptance rate (since last printing): 0.2400
Step 91000, acceptance rate (since last printing): 0.2230
Step 92000, acceptance rate (since last printing): 0.2390
Step 93000, acceptance rate (since last printing): 0.2280
Step 94000, acceptance rate (since last printing): 0.2430
Step 95000, acceptance rate (since last printing): 0.2350
Step 96000, acceptance rate (since last printing): 0.2210
Step 97000, acceptance rate (since last printing): 0.2490
Step 98000, acceptance rate (since last printing): 0.2540
Step 99000, acceptance rate (since last printing): 0.2290
Step 100000, acceptance rate (since last printing): 0.2270
Saving
Saved. It took 0.13 seconds.
results contains 7 items.
baredSC_2d¶
Use the npz content¶
baredSC is mainly used with the command line interface.
By default, only the numpy compressed .npz file is output,
but if the --figure argument is used, it outputs much more.
All the outputs are described in the Outputs page.
We provide an example of a plot using the text output _pdf2d.txt in Customize your baredSC plots.
However, sometimes the user may want to plot information which is not part of the text outputs.
We describe here a script which will use the baredSC_2d output to plot the mean and median on the same plot and
another script which will use more bins in the output to get smoother results.
First we get the value of parameters at each step of MCMC:
In [5]: import numpy as np
...: import baredSC.twod
...: import matplotlib.pyplot as plt
...:
...: output_baredSC = "../example/second_example_2d_cellgroup1_1gauss_nx20.npz"
...:
...: # First option, use the function extract_from_npz
...: mu, cov, ox, oy, oxpdf, oypdf, x, y, \
...: logprob_values, samples = \
...: baredSC.twod.extract_from_npz(output_baredSC)
...: # mu is the mean of each parameter,
...: # cov is the covariance of parameters
...: # ox, oy are the oversampled x, y to compute the poisson noise pdf
...: # oxpdf, oypdf are the oversampled x, y to compute the pdf
...: # x, y are the x, y used to compute the likelihood
...: # logprob_values is the value of log likelihood at each step fo the MCMC
...: # samples is the value of each parameter at each step of the MCMC
...:
...: # Second option, get the samples and x, y, oxpdf, oypdf, samples from the npz:
...: mcmc_res = np.load(output_baredSC, allow_pickle=True)
...: samples = mcmc_res['samples']
...: x = mcmc_res['x']
...: y = mcmc_res['y']
...: oxpdf = mcmc_res['oxpdf']
...: oypdf = mcmc_res['oypdf']
...:
...: # From the sample size we deduce the number of Gaussians
...: nnorm = (samples.shape[1] + 1) // 6
...: # We display the parameter names:
...: p_names = [f'{pn}{i}' for i in range(nnorm)
...: for pn in ['xy_amp', 'xy_mux', 'xy_muy', 'xy_scalex',
...: 'xy_scaley', 'xy_corr']][1:]
...: print(f'The parameters are: {p_names}')
...:
Reading
Read. It took 0.02 seconds.
The parameters are: ['xy_mux0', 'xy_muy0', 'xy_scalex0', 'xy_scaley0', 'xy_corr0']
Then we compute the pdf for each step of the MCMC and we plot the mean and median:
In [6]: # We assume x and y are equally spaced
...: dx = x[1] - x[0]
...: nx = x.size
...: dy = y[1] - y[0]
...: ny = y.size
...: noxpdf = oxpdf.size
...: # We assume oxpdf is equally spaced
...: odxpdf = oxpdf[1] - oxpdf[0]
...:
...: noypdf = oypdf.size
...: # We assume oypdf is equally spaced
...: odypdf = oypdf[1] - oypdf[0]
...:
...: odxypdf = odxpdf * odypdf
...: oxypdf = np.array(np.meshgrid(oxpdf, oypdf)).transpose(1, 2, 0)
...:
...: # Compute the pdf for each sample
...: # This can be long
...: pdf = np.array([baredSC.twod.get_pdf(p, nx, ny, noxpdf,
...: noypdf, oxypdf, odxypdf)
...: for p in samples])
...: # We plot:
...: xmin = x[0] - dx / 2
...: xmax = x[-1] + dx / 2
...: ymin = y[0] - dy / 2
...: ymax = y[-1] + dy / 2
...:
...: x_borders = np.linspace(xmin, xmax, len(x) + 1)
...: y_borders = np.linspace(ymin, ymax, len(y) + 1)
...:
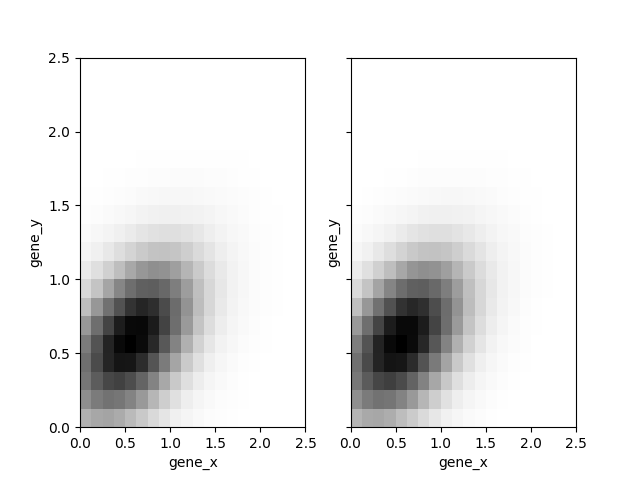
...: # Plot 2 panels plot
...: fig, axs = plt.subplots(1, 2, sharex='row', sharey='row')
...: axs[0].pcolormesh(x_borders, y_borders, np.mean(pdf, axis=0),
...: shading='flat', rasterized=True, cmap='Greys')
...: axs[0].set_xlabel('gene_x')
...: axs[0].set_ylabel('gene_y')
...: axs[1].pcolormesh(x_borders, y_borders, np.median(pdf, axis=0),
...: shading='flat', rasterized=True, cmap='Greys')
...: axs[1].set_xlabel('gene_x')
...: axs[1].set_ylabel('gene_y')
...:
Out[6]: Text(0, 0.5, 'gene_y')
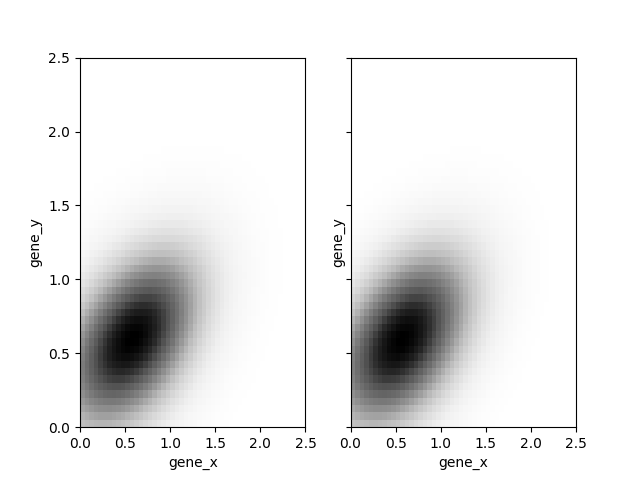
If you want to get more bins, you just need to change x and y. We want to warn the user that what will be plotted will be different from what was used for the likelihood evaluation:
In [1]: # We assume x and y are equally spaced
...: dx = x[1] - x[0]
...: dy = y[1] - y[0]
...: xmin = x[0] - dx / 2
...: xmax = x[-1] + dx / 2
...: ymin = y[0] - dy / 2
...: ymax = y[-1] + dy / 2
...:
...: # We set pretty_bins_x and y
...: pretty_bins_x = 50
...: pretty_bins_y = 50
...: from baredSC.common import get_bins_centers
...: nx = pretty_bins_x
...: x = get_bins_centers(xmin, xmax, nx)
...: dx = x[1] - x[0]
...: noxpdf = nx
...: oxpdf = x
...: odxpdf = dx
...: ny = pretty_bins_y
...: y = get_bins_centers(ymin, ymax, ny)
...: dy = y[1] - y[0]
...: noypdf = ny
...: oypdf = y
...: odypdf = dy
...:
...: odxypdf = odxpdf * odypdf
...: oxypdf = np.array(np.meshgrid(oxpdf, oypdf)).transpose(1, 2, 0)
...:
...: # Compute the pdf for each sample
...: # This can be long
...: pdf = np.array([baredSC.twod.get_pdf(p, nx, ny, noxpdf,
...: noypdf, oxypdf, odxypdf)
...: for p in samples])
...: # We plot:
...: x_borders = np.linspace(xmin, xmax, len(x) + 1)
...: y_borders = np.linspace(ymin, ymax, len(y) + 1)
...:
...:
...: # Plot 2 panels plot
...: fig, axs = plt.subplots(1, 2, sharex='row', sharey='row')
...: axs[0].pcolormesh(x_borders, y_borders, np.mean(pdf, axis=0),
...: shading='flat', rasterized=True, cmap='Greys')
...: axs[0].set_xlabel('gene_x')
...: axs[0].set_ylabel('gene_y')
...: axs[1].pcolormesh(x_borders, y_borders, np.median(pdf, axis=0),
...: shading='flat', rasterized=True, cmap='Greys')
...: axs[1].set_xlabel('gene_x')
...: axs[1].set_ylabel('gene_y')
...:
Run baredSC_2d¶
You can also run the MCMC from python directly.
However, it requires formating of the input:
In [7]: import numpy as np
...: import pandas as pd
...: from scipy.stats import lognorm, truncnorm, poisson
...: from baredSC.baredSC_2d import gauss_mcmc
...: from baredSC.twod import trunc_norm2d
...:
...:
...: def trunc_norm_2d(mu, sigma, corr, size, seed):
...: try:
...: rng = np.random.default_rng(seed)
...: except AttributeError:
...: # For older numpy versions:
...: np.random.seed(seed)
...: rng = np.random
...: cov = np.array([[sigma[0] * sigma[0], sigma[0] * sigma[1] * corr],
...: [sigma[0] * sigma[1] * corr, sigma[1] * sigma[1]]])
...: values = rng.multivariate_normal(mu, cov, size)
...: mask_0 = [v[0] < 0 or v[1] < 0 for v in values]
...: # Because we want only positive expression:
...: while sum(mask_0) > 0:
...: values[mask_0] = rng.multivariate_normal(mu, cov, sum(mask_0))
...: mask_0 = [v[0] < 0 or v[1] < 0 for v in values]
...: return(values)
...:
...:
...: # I generate 200 cells with normal expression at 1.5 with scale of 0.2 with correlation of 0.5
...: # In the Seurat scale (log(1 + 10^4 X))
...: n_cells = 200
...: cur_mu = [1.5, 1.5]
...: cur_sigma = [0.2, 0.2]
...: cur_corr = 0.5
...: N = lognorm.rvs(s=0.3, scale=16000, size=n_cells, random_state=1).astype(int)
...: expression = trunc_norm_2d(mu=cur_mu, sigma=cur_sigma,
...: corr=cur_corr,
...: size=n_cells,
...: seed=2)
...: exp_values_x, exp_values_y = np.transpose(expression)
...: ks_x = poisson.rvs(mu=N * 1e-4 * (np.exp(exp_values_x) - 1),
...: random_state=3)
...: ks_y = poisson.rvs(mu=N * 1e-4 * (np.exp(exp_values_y) - 1),
...: random_state=4)
...:
...: # I need to put the ks and the N in a data frame:
...: # The column containing the total number of UMI per cell
...: # must be 'nCount_RNA'
...: data = pd.DataFrame({'my_gene_x': ks_x,
...: 'my_gene_y': ks_y,
...: 'nCount_RNA': N})
...:
Then the actual MCMC can be run with:
In [8]: results = gauss_mcmc(data=data,
...: genex='my_gene_x', # Put here the colname you put in your data
...: geney='my_gene_y', # Put here the colname you put in your data
...: nx=20, # Number of bins in x
...: osampx=10, # Oversampling factor of the Poisson distribution
...: osampxpdf=5, # Oversampling factor of the PDF
...: xmin=0,
...: xmax=3,
...: ny=20, # Number of bins in y
...: osampy=10, # Oversampling factor of the Poisson distribution
...: osampypdf=5, # Oversampling factor of the PDF
...: ymin=0,
...: ymax=3,
...: min_scale_x=0.1, # Minimal value of the scale in x
...: min_scale_y=0.1, # Minimal value of the scale in y
...: scale_prior=0.3, # Scale of the truncnorm used in the prior for the correlation
...: scale="Seurat",
...: target_sum=10000,
...: nnorm=1, # We use models with a single Gaussian
...: nsamples_mcmc=100000, # Number of steps in the MCMC
...: nsamples_burn=25000, # Number of steps in the burning phase of MCMC (we recommand nsampMCMC / 4)
...: nsplit_burn=10, # The burning phase is splitted in multiple sub-phase where the temperature is decreasing
...: T0_burn=100.0,
...: output='temp', # Where the npz output should be stored
...: seed=1)
...: print(f'results contains {len(results)} items.')
...: # The results are:
...: # mu, cov, ox, oy, oxpdf, oypdf, x, y, \
...: # logprob_values, samples
...: # mu is the mean of each parameter,
...: # cov is the covariance of parameters
...: # ox, oy are the oversampled x, y to compute the poisson noise pdf
...: # oxpdf, oypdf are the oversampled x, y to compute the pdf
...: # x, y are the x, y used to compute the likelihood
...: # logprob_values is the value of log likelihood at each step fo the MCMC
...: # samples is the value of each parameter at each step of the MCMC
...:
Step 1000, acceptance rate (since last printing): 0.2320
Step 2000, acceptance rate (since last printing): 0.2180
Step 1000, acceptance rate (since last printing): 0.2710
Step 2000, acceptance rate (since last printing): 0.2270
Step 1000, acceptance rate (since last printing): 0.2670
Step 2000, acceptance rate (since last printing): 0.1990
Step 1000, acceptance rate (since last printing): 0.2650
Step 2000, acceptance rate (since last printing): 0.2220
Step 1000, acceptance rate (since last printing): 0.3320
Step 2000, acceptance rate (since last printing): 0.1970
Step 1000, acceptance rate (since last printing): 0.2010
Step 2000, acceptance rate (since last printing): 0.2160
Step 1000, acceptance rate (since last printing): 0.2570
Step 2000, acceptance rate (since last printing): 0.2210
Step 1000, acceptance rate (since last printing): 0.2650
Step 2000, acceptance rate (since last printing): 0.2300
Step 1000, acceptance rate (since last printing): 0.2410
Step 2000, acceptance rate (since last printing): 0.2590
Step 1000, acceptance rate (since last printing): 0.2520
Step 2000, acceptance rate (since last printing): 0.2280
Step 1000, acceptance rate (since last printing): 0.2490
Step 2000, acceptance rate (since last printing): 0.2170
Step 3000, acceptance rate (since last printing): 0.2480
Step 4000, acceptance rate (since last printing): 0.2440
Step 5000, acceptance rate (since last printing): 0.2110
Step 6000, acceptance rate (since last printing): 0.2280
Step 7000, acceptance rate (since last printing): 0.2620
Step 8000, acceptance rate (since last printing): 0.2480
Step 9000, acceptance rate (since last printing): 0.2350
Step 10000, acceptance rate (since last printing): 0.2210
Step 11000, acceptance rate (since last printing): 0.2130
Step 12000, acceptance rate (since last printing): 0.2540
Step 13000, acceptance rate (since last printing): 0.2420
Step 14000, acceptance rate (since last printing): 0.2330
Step 15000, acceptance rate (since last printing): 0.2300
Step 16000, acceptance rate (since last printing): 0.2430
Step 17000, acceptance rate (since last printing): 0.2440
Step 18000, acceptance rate (since last printing): 0.2470
Step 19000, acceptance rate (since last printing): 0.2230
Step 20000, acceptance rate (since last printing): 0.2260
Step 21000, acceptance rate (since last printing): 0.2460
Step 22000, acceptance rate (since last printing): 0.2310
Step 23000, acceptance rate (since last printing): 0.2400
Step 24000, acceptance rate (since last printing): 0.2580
Step 25000, acceptance rate (since last printing): 0.2510
Step 26000, acceptance rate (since last printing): 0.2530
Step 27000, acceptance rate (since last printing): 0.2200
Step 28000, acceptance rate (since last printing): 0.2520
Step 29000, acceptance rate (since last printing): 0.2380
Step 30000, acceptance rate (since last printing): 0.2460
Step 31000, acceptance rate (since last printing): 0.2560
Step 32000, acceptance rate (since last printing): 0.2170
Step 33000, acceptance rate (since last printing): 0.2500
Step 34000, acceptance rate (since last printing): 0.2540
Step 35000, acceptance rate (since last printing): 0.2340
Step 36000, acceptance rate (since last printing): 0.2390
Step 37000, acceptance rate (since last printing): 0.2450
Step 38000, acceptance rate (since last printing): 0.2440
Step 39000, acceptance rate (since last printing): 0.2410
Step 40000, acceptance rate (since last printing): 0.2480
Step 41000, acceptance rate (since last printing): 0.2160
Step 42000, acceptance rate (since last printing): 0.2420
Step 43000, acceptance rate (since last printing): 0.2500
Step 44000, acceptance rate (since last printing): 0.2280
Step 45000, acceptance rate (since last printing): 0.2300
Step 46000, acceptance rate (since last printing): 0.2540
Step 47000, acceptance rate (since last printing): 0.2120
Step 48000, acceptance rate (since last printing): 0.2340
Step 49000, acceptance rate (since last printing): 0.2380
Step 50000, acceptance rate (since last printing): 0.2460
Step 51000, acceptance rate (since last printing): 0.2500
Step 52000, acceptance rate (since last printing): 0.2440
Step 53000, acceptance rate (since last printing): 0.2310
Step 54000, acceptance rate (since last printing): 0.2330
Step 55000, acceptance rate (since last printing): 0.2340
Step 56000, acceptance rate (since last printing): 0.2320
Step 57000, acceptance rate (since last printing): 0.2440
Step 58000, acceptance rate (since last printing): 0.2560
Step 59000, acceptance rate (since last printing): 0.2360
Step 60000, acceptance rate (since last printing): 0.2550
Step 61000, acceptance rate (since last printing): 0.2240
Step 62000, acceptance rate (since last printing): 0.2500
Step 63000, acceptance rate (since last printing): 0.2190
Step 64000, acceptance rate (since last printing): 0.2370
Step 65000, acceptance rate (since last printing): 0.2200
Step 66000, acceptance rate (since last printing): 0.2170
Step 67000, acceptance rate (since last printing): 0.2250
Step 68000, acceptance rate (since last printing): 0.2400
Step 69000, acceptance rate (since last printing): 0.2520
Step 70000, acceptance rate (since last printing): 0.2780
Step 71000, acceptance rate (since last printing): 0.2220
Step 72000, acceptance rate (since last printing): 0.2310
Step 73000, acceptance rate (since last printing): 0.2450
Step 74000, acceptance rate (since last printing): 0.2380
Step 75000, acceptance rate (since last printing): 0.2190
Step 76000, acceptance rate (since last printing): 0.2470
Step 77000, acceptance rate (since last printing): 0.2450
Step 78000, acceptance rate (since last printing): 0.2320
Step 79000, acceptance rate (since last printing): 0.2380
Step 80000, acceptance rate (since last printing): 0.2530
Step 81000, acceptance rate (since last printing): 0.2330
Step 82000, acceptance rate (since last printing): 0.2370
Step 83000, acceptance rate (since last printing): 0.2360
Step 84000, acceptance rate (since last printing): 0.2310
Step 85000, acceptance rate (since last printing): 0.2290
Step 86000, acceptance rate (since last printing): 0.2660
Step 87000, acceptance rate (since last printing): 0.2400
Step 88000, acceptance rate (since last printing): 0.2110
Step 89000, acceptance rate (since last printing): 0.2380
Step 90000, acceptance rate (since last printing): 0.2530
Step 91000, acceptance rate (since last printing): 0.2200
Step 92000, acceptance rate (since last printing): 0.2400
Step 93000, acceptance rate (since last printing): 0.2170
Step 94000, acceptance rate (since last printing): 0.2320
Step 95000, acceptance rate (since last printing): 0.2320
Step 96000, acceptance rate (since last printing): 0.2260
Step 97000, acceptance rate (since last printing): 0.2440
Step 98000, acceptance rate (since last printing): 0.2390
Step 99000, acceptance rate (since last printing): 0.2790
Step 100000, acceptance rate (since last printing): 0.2310
Saving
Saved. It took 0.18 seconds.
results contains 10 items.